In this session, we will perform reference-anchored modification calling
on the dataset we have been using thus far — yeast DNA where some thymidines have been
substituted by BrdU.
We will use the software program DNAscent on the files already generated
by us from previous sessions (fast5, bam, sequencing summary file).
We will then convert the output of DNAscent into the standard mod bam format for storing
modification information.
Along the way, we will also learn the mod bam file format.
We will be performing the steps highlighted with an asterisk in the pipeline figure below.
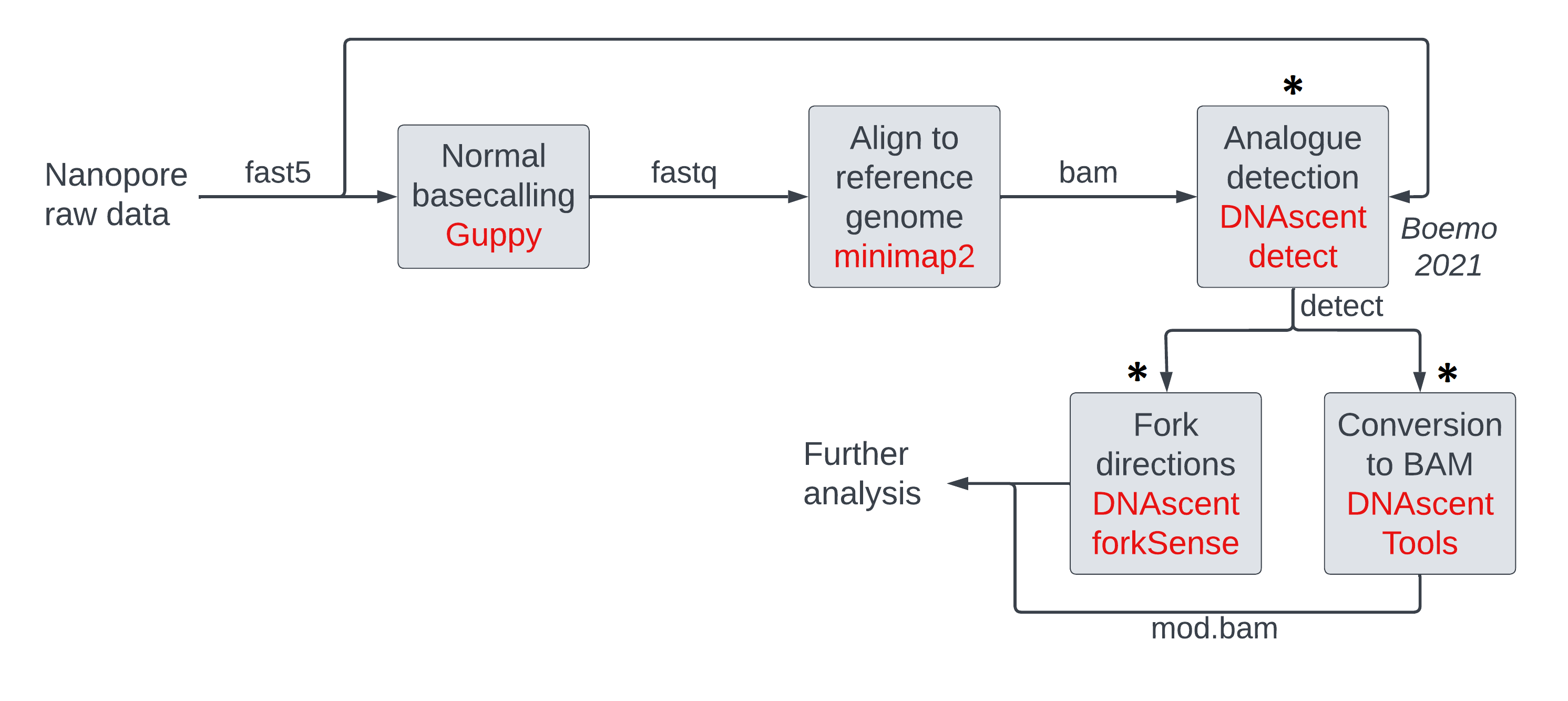
Running DNAscent to produce modification calls along each sequenced DNA strand
Optional: downloading a plugin required by DNAscent
Preparations to call modifications
We first need to download a plugin to help DNAscent read the later versions of fast5 files.
mkdir -p ~/nanomod_course_references # any suitable folder will do here.
cd ~/nanomod_course_references
wget https://github.com/nanoporetech/vbz_compression/releases/download/v1.0.1/ont-vbz-hdf-plugin-1.0.1-Linux-x86_64.tar.gz
tar -xvzf ont-vbz-hdf-plugin-1.0.1-Linux-x86_64.tar.gz
export HDF5_PLUGIN_PATH=$(pwd)/ont-vbz-hdf-plugin-1.0.1-Linux/usr/local/hdf5/lib/plugin
ls $HDF5_PLUGIN_PATH
# libvbz_hdf_plugin.so
rm ont-vbz-hdf-plugin-1.0.1-Linux-x86_64.tar.gz # if you see the output above, then cleanup by removing the tarball.
Creating a DNAscent index
We now need to create an index needed by DNAscent — a plain text with two columns: read id and the fast5 file with the corresponding time course of nanopore current.
input_fast5_dir=~/nanomod_course_data/yeast
output_index=~/nanomod_course_outputs/yeast/index.dnascent
input_seq_summ=~/nanomod_course_outputs/yeast/sequencing_summary.txt
DNAscent index -f $input_fast5_dir -o $output_index \
-s $input_seq_summ
Call modifications
We run the reference-anchored modification caller DNAscent detect here.
The program takes alignment information from the bam file, a reference genome,
and the raw nanopore currents as inputs and outputs the probability of modification
per thymidine per sequenced strand in a .detect format,
which is very similar to the TSV (tab-separated values) format.
The program uses a machine-learning approach where model parameters learned previously
represent differences in nanopore current characteristics between thymidine and BrdU.
The additional options below direct the program to use
8 computational threads and to reject alignments below a mapping quality of 20 and
a genome-mapped length of 1000 bases.
By omitting the --GPU input parameter, we are running DNAscent in a slow, CPU-only mode
as the virtual machines used in the course do not have GPUs to lower costs.
input_bam=~/nanomod_course_outputs/yeast/aligned_reads.sorted.onlyPrim.bam
ref_genome=~/nanomod_course_references/sacCer3.fa
index=~/nanomod_course_outputs/yeast/index.dnascent
output_detect=~/nanomod_course_outputs/yeast/dnascent.detect
DNAscent detect -b $input_bam -r $ref_genome -i $index \
-o $output_detect -t 8 -q 20 -l 1000
After the program has finished running, inspect the first few lines of the output
file using the head command
output_detect=~/nanomod_course_outputs/yeast/dnascent.detect
head -n 30 $output_detect
You should see:
- a few header lines starting with
# - data from each read starts with
>and has information about its alignment - the following lines give genomic alignment coordinate, BrdU probability, and k-mer (on the reference strand)
Call replication dynamics with DNAscent forkSense using single-molecule modification densities
After modifications are called, one can ask general questions common to all modification experiments like which are the highly modified reads, what is the mean density per read etc. One can also ask specific questions that are experiment-dependent.
In the experiments of the yeast dataset we have been using,
the specific questions are about DNA replication dynamics.
A specialized program called forkSense that ships with DNAscent detects gradients in BrdU density
across a read, and associates these with the movement of replication forks.
After fork calls are made, locations on the genome from which forks emerged are marked as
origins of replication, and locations where forks terminate are marked as termination sites.
Please see the figure below, reproduced from Müller et al., Nat. Methods (2019) at
this link.
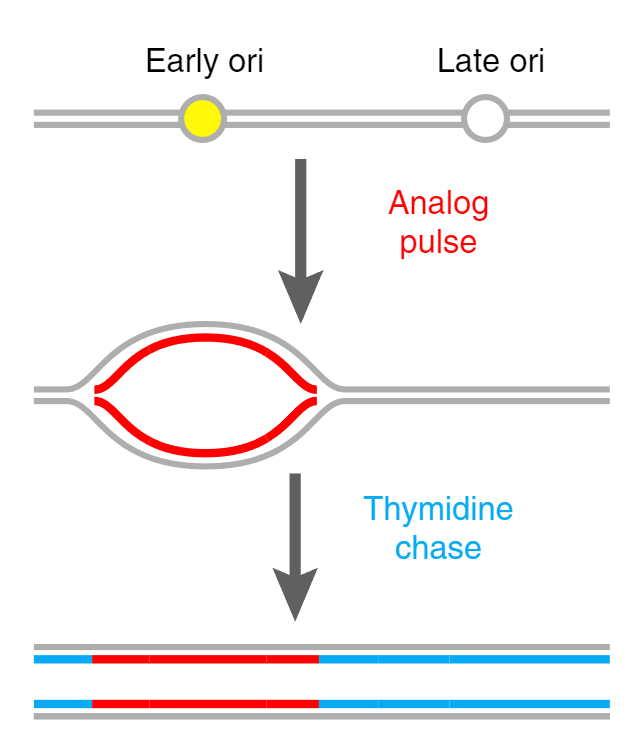
In the course, we will just learn how to execute the forkSense command.
Please discuss the biological interpretations with the instructors if you are interested.
# Please change to the output directory using the cd command.
# There are lots of output files which go to the output directory.
input_detect=~/nanomod_course_outputs/yeast/dnascent.detect
output_forksense=~/nanomod_course_outputs/yeast/dnascent.forkSense
cd ~/nanomod_course_outputs/yeast/
DNAscent forkSense -d $input_detect -o $output_forksense \
-t 8 --markOrigins --markTerminations --markForks
The above command will produce a few plain-text output files.
Inspect them using shuf and head; their names should be self-explanatory.
Conversion of DNAscent detect into the modBAM format
We convert modification data from the specific .detect format used by DNAscent to
the more widely used and standardized .mod.bam or mod BAM format using the package
DNAscentTools developed by the Nieduszynski group.
Mod BAM files are just BAM files with two additional tags per line that store
modification information. We will discuss their structure later on in this session.
input_detect=~/nanomod_course_outputs/yeast/dnascent.detect
output_mod_bam=~/nanomod_course_outputs/yeast/dnascent.detect.mod.bam
cd ~/nanomod_course_scripts/DNAscentTools
< $input_detect python convert_detect_to_modBAM.py \
--op $output_mod_bam --tag T
# sort and index the BAM files
input_mod_bam=~/nanomod_course_outputs/yeast/dnascent.detect.mod.bam
output_mod_bam=~/nanomod_course_outputs/yeast/dnascent.detect.mod.sorted.bam
samtools sort -o $output_mod_bam $input_mod_bam
samtools index $output_mod_bam
Inspect modification data by conversion from modBAM to TSV format using modkit
Without a discussion, it is not easy to understand what is in a raw mod BAM file.
So, before the discussion, let us convert the mod BAM file into the easy-to-understand tab-separated value
format so that we can get a quick look at our modification calls.
We will be using the modkit program developed by ONT that takes mod BAM files as input
and produces tabulated data or summary statistics as output.
One of the functions provided is modkit extract.
Please run the code below and inspect the output tsv file.
The most important columns are read_id, forward_read_position, and mod_qual as these answer
the fundamental question of modification calling: what is the likelihood of modification at every
position on every read?
Other useful columns are the position along the reference ref_position,
the length of the read read_length etc.
We can look up the full list in the official documentation here.
input_mod_bam=~/nanomod_course_outputs/yeast/dnascent.detect.mod.sorted.bam
output_tsv=~/nanomod_course_outputs/yeast/dnascent.detect.mod.sorted.bam.tsv
modkit extract $input_mod_bam $output_tsv
Optional: the modBAM format
Discussion of the modBAM file format
The mod BAM format is not very easy to read for a person.
Tools such as modkit, samtools, and IGV help us convert mod BAM files into an easily
readable format such as a table or an image.
You might find that such tools are adequate for your needs.
If you do not, then knowledge of the format helps you write tools yourself
and to perform some advanced manipulations.
As the modification field is still developing,
it is probable that you will come across such scenarios.
Thus, we conclude this session with a discussion
of the mod BAM file format.